Abstract
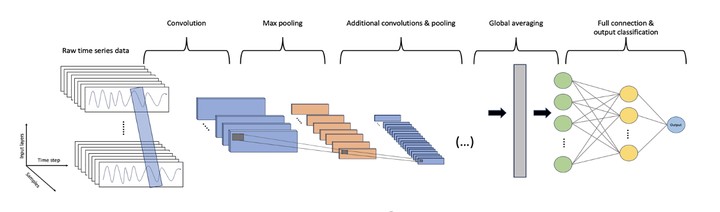
Species distribution models (SDMs) have become indispensable tools for relating species occurrences to environmental conditions. Most commonly, these models utilize machine learning algorithms to statistically evaluate environmental covariates and determine the relative suitability of locations on the landscape. Currently these models are limited to static predictors, usually constructed from averaging climate data data over extended periods. However, almost all factors driving species distributions are temporally dynamic. That is, their state changes with time, a property often poorly represented, or entirely missing, from SDMs. A more subtle, common omission in predictors used in SDMs concerns with the order in which events take place. One way to robustly address these limitations is by calibrating SDMS that consider the full representation pf spatial and temporal variability in predictor sets. Recent architectures of deep learning neural networks allow dealing with fully explicit spatiotemporal dynamics, thus fitting SDMs without the need to simplify temporal and spatial dimensions of predictor data. We present a deep learning based SDM approach that uses time series of spatial data for a variety of taxa and compare these models to conventional methods. Deep learning approaches consistently provided high performing models while avoiding the use of pre-processed predictor sets that can obscure relevant aspects if environmental variation.